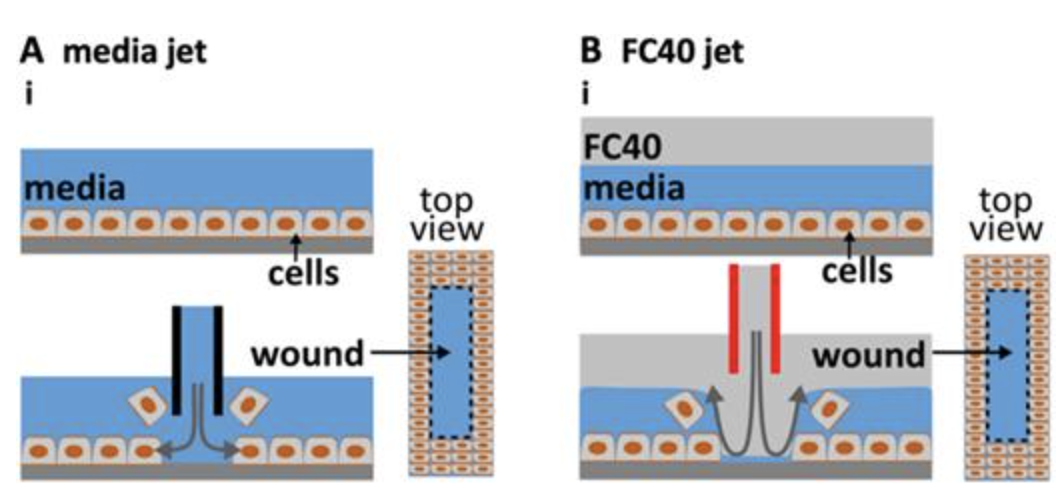
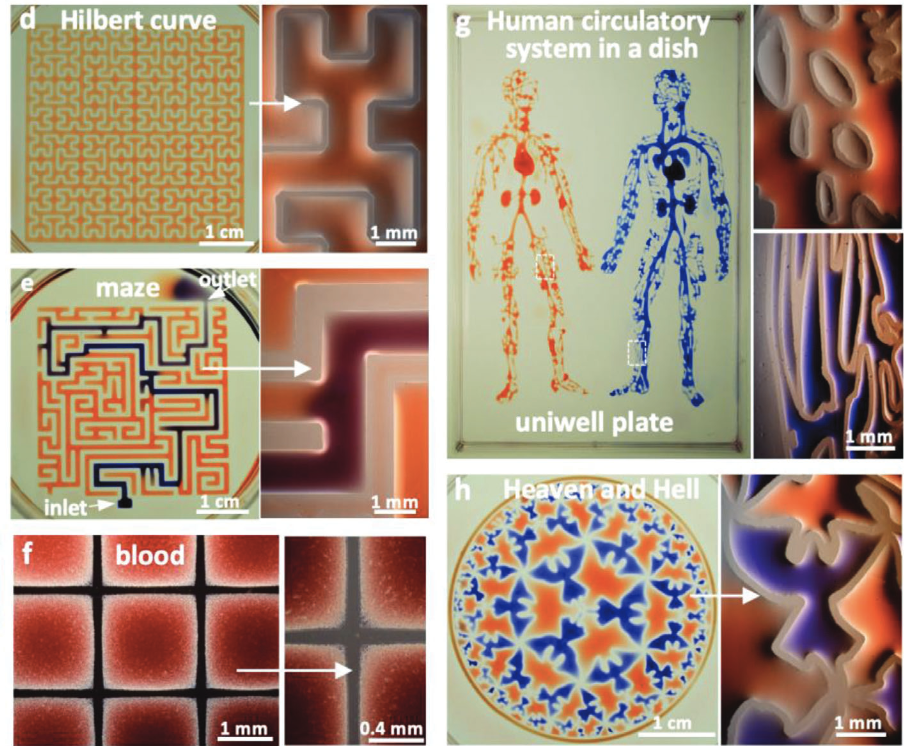
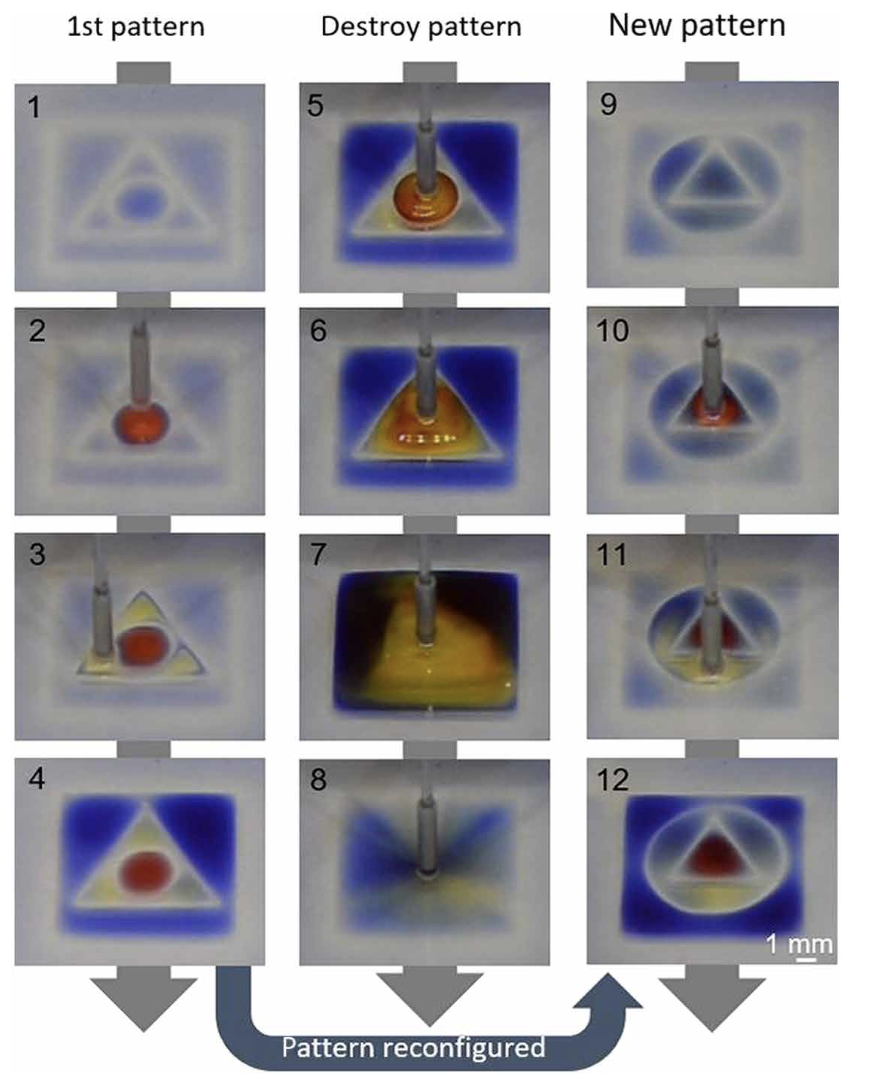
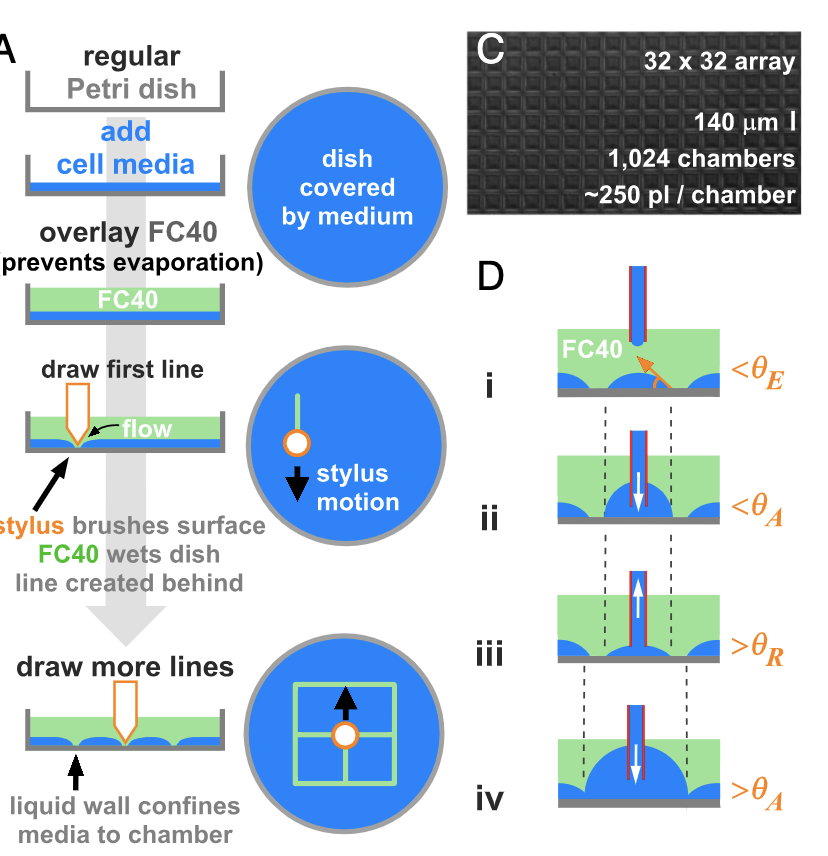
Cristian Soitu
Computational Biology Researcher & ML/AI Scientist
An ML Engineer with a strong background in developing and applying machine learning models to complex biological data. Experienced in the end-to-end development of computational tools for 3D bio-imaging and multi-modal data analysis. A proficient Python developer focused on leveraging advanced AI techniques, including Transformers and CNNs, to solve challenges in oncology and neuroscience.